Zhaoxiang (Simon) Cai
Senior Data Scientist
The University of Sydney
Children's Medical Research Institute
Biography
I am a dynamic researcher and engineer with a robust background in both academic and industrial settings. My journey includes developing large-scale systems at Goldman Sachs, where I successfully managed complex projects under tight deadlines. My PhD in Cancer Data Science at the Children’s Medical Research Institute, affiliated with the University of Sydney, marked a significant turn in my career towards integrating machine learning with oncological studies. My passion lies in harnessing the power of artificial intelligence to revolutionise healthcare, particularly in understanding and treating cancer. I am committed to pioneering advancements that will transform oncology, speeding the development of innovative therapies and improving patient outcomes. Beyond my professional pursuits, I am an avid enthusiast of the piano, skiing, and badminton.
Download my resumé .
- Artificial Intelligence
- Cancer Research
- Big Data
- Computer Vision
PhD in Machine Learning and Medical Research, 2023
The University of Sydney
Master in Business Analytics, 2018
The University of Melbourne
Hons of BSc in Computer Science, 2014
Monash University
Skills
Experience
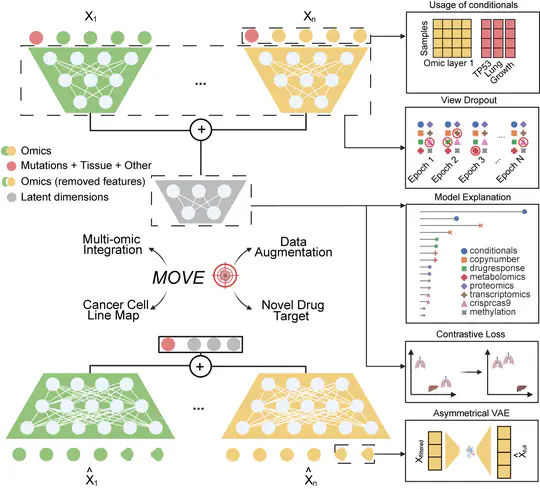
- Developed new deep learning-based approach to incorporate human knowledge for multi-omic data integration
- Designed and built multi-view VAE models customised for multi-omic data integration
- Performed end-to-end whole exome/genome sequencing data analyses for germline/somatic mutations, copy number variations and structural variants
- Performed end-to-end proteomic data analyses, including data QC, peptide-to-protein rollup, pre-processing, differential expression analysis, pathway analysis and survival analysis
- Worked on integrating histopathological images with proteomic data to improve diagnosis
- Fundamentals of Programming
- Software Engineering with Java
- Built pipelines using existing models for single-cell RNA-seq analysis in mouse developmental biology
- Built deep learning models for live-cell imaging data analysis
- Communicated with business stakeholders and liaised regarding project scope with ongoing updates
- Designed/developed/tested/deployed system solutions specialised in Goldman Sachs Electronic Trading (GSET)business flow
- Provided production support and maitained the health of testing environment
Featured Publications
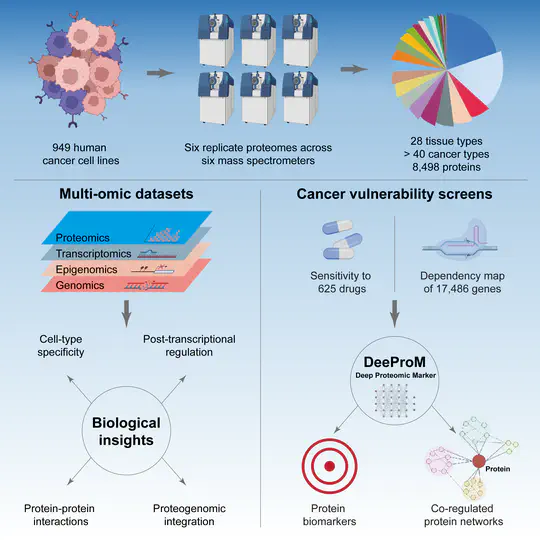
The proteome provides unique insights into disease biology beyond the genome and transcriptome. A lack of large proteomic datasets has restricted the identification of new cancer biomarkers. Here, proteomes of 949 cancer cell lines across 28 tissue types are analyzed by mass spectrometry. Deploying a workflow to quantify 8,498 proteins, these data capture evidence of cell-type and post-transcriptional modifications. Integrating multi-omics, drug response, and CRISPR-Cas9 gene essentiality screens with a deep learning-based pipeline reveals thousands of protein biomarkers of cancer vulnerabilities that are not significant at the transcript level. The power of the proteome to predict drug response is very similar to that of the transcriptome. Further, random downsampling to only 1,500 proteins has limited impact on predictive power, consistent with protein networks being highly connected and co-regulated. This pan-cancer proteomic map (ProCan-DepMapSanger) is a comprehensive resource available at https://cellmodelpassports.sanger.ac.uk.